Final Technical Report - Analysis of the molecular profile of vaccines

Preface
First of all we would like to thank the very useful comments provided by those who have viewed the results of the analyzes, carried out in the context of scientific research activities, relating to the Priorix tetra and Infanrix Hexa products. The critical issues exposed were in fact very useful in order to add technical-scientific integrations capable of clarifying the work done. We believe that only through a healthy communion of scientific visions can conclusions be reached on the data obtained that can be useful for the entire scientific community and for the people who turn to it.
1. State of the art
The preliminary studies (screening not subject to confirmation) of biomolecular, metabolomic and proteomic profile, performed on the Priorix Tetra and Infanrix Hexa products have led to a composition composition summarized in the following points:
- Presence of different analytical signals that cannot be associated with known compounds through searches on Metlin databases 1-2 and KEGG 3. Therefore, a picture emerged associated with a considerable complexity in the composition of commercial products.
- Presence of undeclared proteins in the leaflet in the Priorix Tetra product. The latter can potentially be associated with residues from the production process
- Non detection of the antigens declared inside the Infanrix Hexa product. The analysis technique consisted of enzymatic digestion with trypsin associated with mass spectrometry techniques. 4-5
These data raised several comments, especially regarding point C - The detection of proteins is in fact carried out using a standard approach, recognized internationally for over 10 years 4digestion through the trypsin enzyme 4. The peptides thus obtained are separated chromatographically and analyzed by mass spectrometry 4-5. The main observation was inherent in the fact that aluminum-based adjuvant compounds are present in vaccines that could potentially inhibit the enzymatic digestion process.
The data acquired subsequently made it possible to provide substantive clarifications, especially regarding the dispute found in point c.
2. New insights and analyzes
2.1 Further information on the analysis of the Infanrix Hexa product
Before proceeding with the illustration of the new acquired data concerning the Hexyon and Gardasil 9 vaccines, it is imperative to answer the question inherent in the doubt raised regarding the inhibition of the proteolytic activity of trypsin caused by the presence of aluminum-based adjuvants. in the Infanrix Hexa vaccine. In this regard, it must be specified that a digestion control is always present within the tryptic digestate. In fact, the trypsin used to carry out digestion, although engineered for the purpose of preventing autolysis, has a small percentage of the latter which in case of enzymatic activity leads to the obtainment of the fragment having m / z 842 and the following peptide sequence : VATVSLPR. This fragment was actually detected inside the tryptic digestate of the Infanrix Hexa product as verifiable by the ion extraction chromatogram (Figure 1).
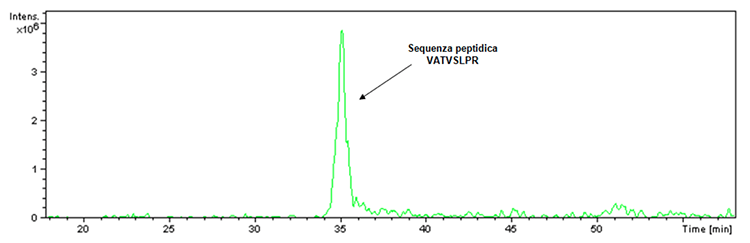
Figure 1: Ion extraction chromatogram associated with the ion at m / z ratio 842 detected in the sample relating to the batch of the Infanrix Hexa product (batch number A21CD072D).
In addition, an external check is performed by digesting hemoglobin, in order to further verify the goodness of the lot of trypsin used. Hemoglobin, analyzed in the analysis section in which the product was monitored, was recognized with a significant statistical score (loge <-100). These data allowed to confirm the fact that the enzyme activity was present.
2.2 New analyzes related to Hexyon and Gardasil products
The analysis of the Hexyon and Gardasil 9 products led to the detection of complex molecular profiles. In this case, however, the presence of most of the antigens reported in the leaflet was detected. They were detected by operating by tryptic digestion and in the presence of adjuvants. This fact further reinforces the evidence that the tryptic digestion reaction is not inhibited in the presence of adjuvants.
In the case of the Hexyon and Gardasil 9 vaccines, the complexity of the molecular profile was mainly attributed to the presence of numerous species, with low molecular weight, not identifiable through the Metlin reference databases 1-2 and KEGG 3.
3. Conclusions and final considerations
The analyzes carried out led to the following conclusion:
- The molecular profile of the vaccines analyzed is generally complex and largely unknown.
- There are protein contaminations, not declared in the leaflet, whose composition is variable.
- In several cases the antigens declared in the leaflet were not detected. This fact could be attributed to several factors. Among the latter, we can consider the sensitivity of the method used. However, we feel we can exclude the phenomenon of inhibition of digestion due to the presence of adjuvants in the formulation of the vaccine. In fact, the enzymatic activity is confirmed mainly by the presence of tryptic autolysis fragments, inside the solutions of the digested vaccines (internal control).
4. Future studies
Further studies will be carried out as part of the research and development activities aimed at investigating the following aspects:
- macromolecular composition associated with solid residues present in vaccines (MALDI TOF MS analysis); 5
- evaluation of the concentration of metals present in the products.
- Second level analysis to confirm the presence of toxic compounds detected in the screening phase. Their concentration will therefore be related to their toxicity based on what is stated in the international safety data sheets. The second level analyzes will be carried out in compliance with the European directive EU 2002/657 / EC useful to guarantee high quality standards in the mass spectrometry sector. 6
in faith
Dr Loretta Bolgan
5. Bibliographic references
- Autenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti G, Siuzdak G (September 2012). "An accelerated workflow for untargeted metabolomics using the METLIN database". Nature Biotechnology. 30: 826–828. doi: 10.1038 / nbt.2348. PMC 3666346. PMID 22965049.
- Smith CA, I'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (December 2005). "METLIN: a metabolite mass spectral database" (PDF). Ther Drug Monit. 27 (6): 747–51. doi: 10.1097 / 01.ftd.0000179845.53213.39. PMID 16404815.
- Kanehisa M (2013). "Chemical and genomic evolution of enzyme-catalyzed reaction networks". FEBS Lett. 587 (17): 2731–7.
- Cristoni S, Bernardi LR. "Bioinformatics in mass spectrometry data analysis for proteomics studies." Expert Rev Proteomics. 2004 Dec; 1 (4): 469-83.
- Cristoni S, Bernardi LR. "Development of new methodologies for the mass spectrometry study of bioorganic macromolecules." Mass Spectrom Rev. 2003 Nov-Dec; 22 (6): 369-406.
- Cristoni S, Dusi G, Brambilla P, Albini A, Conti M, Brambilla M, Bruno A, Di Gaudio F, Ferlin L, Tazzari V, Mengozzi S, Barera S, Sialer C, Trenti T, Cantu M, Rossi Bernardi L, Noonan DM. "SANIST: optimization of a technology for compound identification based on the European Union directive with applications in forensic, pharmaceutical and food analyzes." J. Mass Spectrom. 2017 Jan; 52 (1): 16-21. doi: 10.1002 / jms.3895.
Download: CORVELVA-Report-technique-Finale.pdf

